ISBI 2013 challenge: 3D segmentation of neurites in EM images
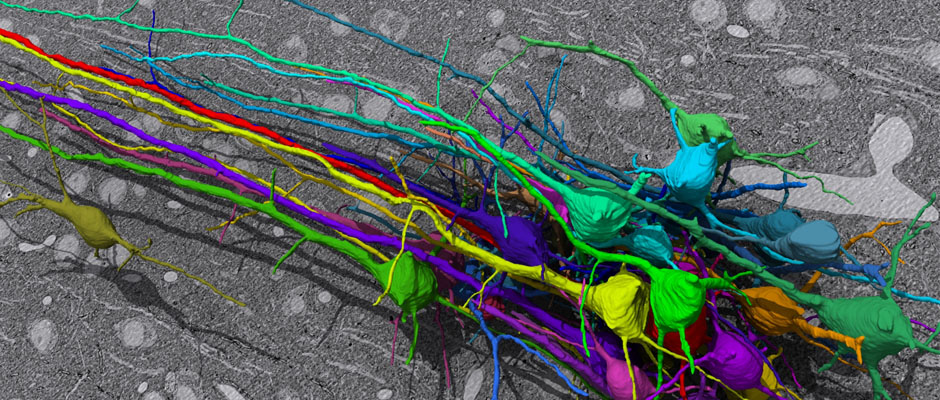 Group of pyramidal neurons of mouse cortex reconstructed from SEM images (Daniel Berger). Group of pyramidal neurons of mouse cortex reconstructed from SEM images (Daniel Berger).
|
Welcome to the server of the first challenge on 3D segmentation of neurites in EM images!
The challenge is organized in the context of the IEEE International Symposium on Biomedical Imaging (San Francisco, CA, April 7-11th 2013). If you wish to participate, please register now to be able to download the training and test data sets and upload your own results.
NEW: for logistic reasons, there is a limit of 10 submissions per day for each user.
Background and set-up
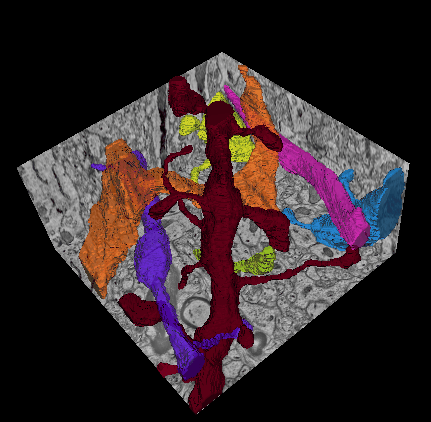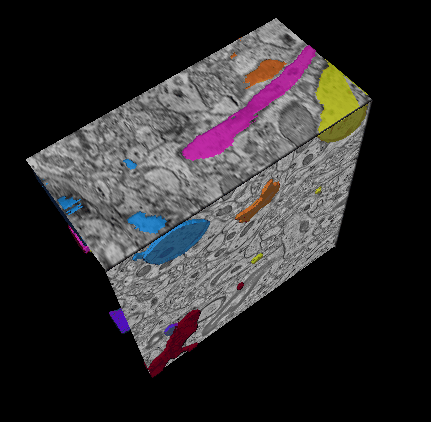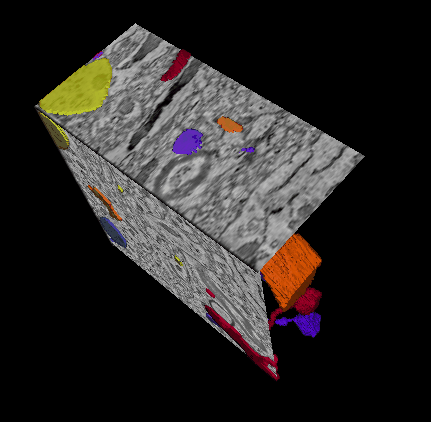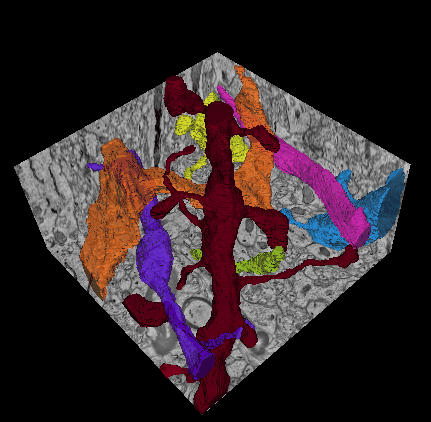
In this challenge, a full stack of electron microscopy (EM) slices will be used to train machine-learning algorithms for the purpose of automatic segmentation of neurites in 3D. This imaging technique visualizes the resulting volumes in a highly anisotropic way, i.e., the x- and y-directions have a high resolution, whereas the z-direction has a low resolution, primarily dependent on the precision of serial cutting. EM produces the images as a projection of the whole section, so some of the neural membranes that are not orthogonal to a cutting plane can appear very blurred. None of these problems led to major difficulties in the manual labeling of each neurite in the image stack by an expert human neuro-anatomist.
In order to gauge the current state-of-the-art in automated neurite segmentation on EM and compare between different methods, we are organizing a 3D Segmentation of neurites in EM images (SNEMI3D) challenge in conjunction with the ISBI 2013 conference. For this purpose, we are making available a large training dataset of mouse cortex in which the neurites have been manually delineated. In addition, we also provide a test dataset where the 3D labels are not available. The aim of the challenge is to compare and rank the different competing methods based on their object classification accuracy in three dimensions.
The image data used in the challenge was produced by Lichtman Lab at Harvard University (Daniel R. Berger, Richard Schalek, Narayanan "Bobby" Kasthuri, Juan-Carlos Tapia, Kenneth Hayworth, Jeff W. Lichtman) and manually annotated by Daniel R. Berger. Their corresponding biological findings were published in Cell (2015).
|
|
Challenge format
Important dates
- January 15th, 2013: training and (first) test data sets released.
- March 30th, 2013: deadline for submitting progress test results and method abstract.
- March 31th, 2013: progress ranking based on first test set and notification of acceptance/presentation type.
- April 7th, 2013: method presentations at the challenge workshop (ISBI 2013 conference).
- TBA: second and final test set released.
- TBA: final deadline test results and method abstract.
- TBA: final ranking based on second dataset.
Results format
The results are expected to be uploaded in the challenge server as a 16-bit TIFF 3D image compressed in a ZIP file. Pixels with the same value (ID) are assume to belong to the same object in 3D. This is exactly the same format as the training labels.